Single-Cell Analysis of Macrophage Abundance and Function in Pancreatic Ductal Adenocarcinoma: Implications for Tumor Progression and Therapy
Code: G-1509
Authors: Hediyeh Khashei ℗, Modjtaba Emadi-Baygi *, Maryam Lotfi-Shahreza
Schedule: Not Scheduled!
Tag: System Biology
Download: Download Poster
Abstract:
Abstract
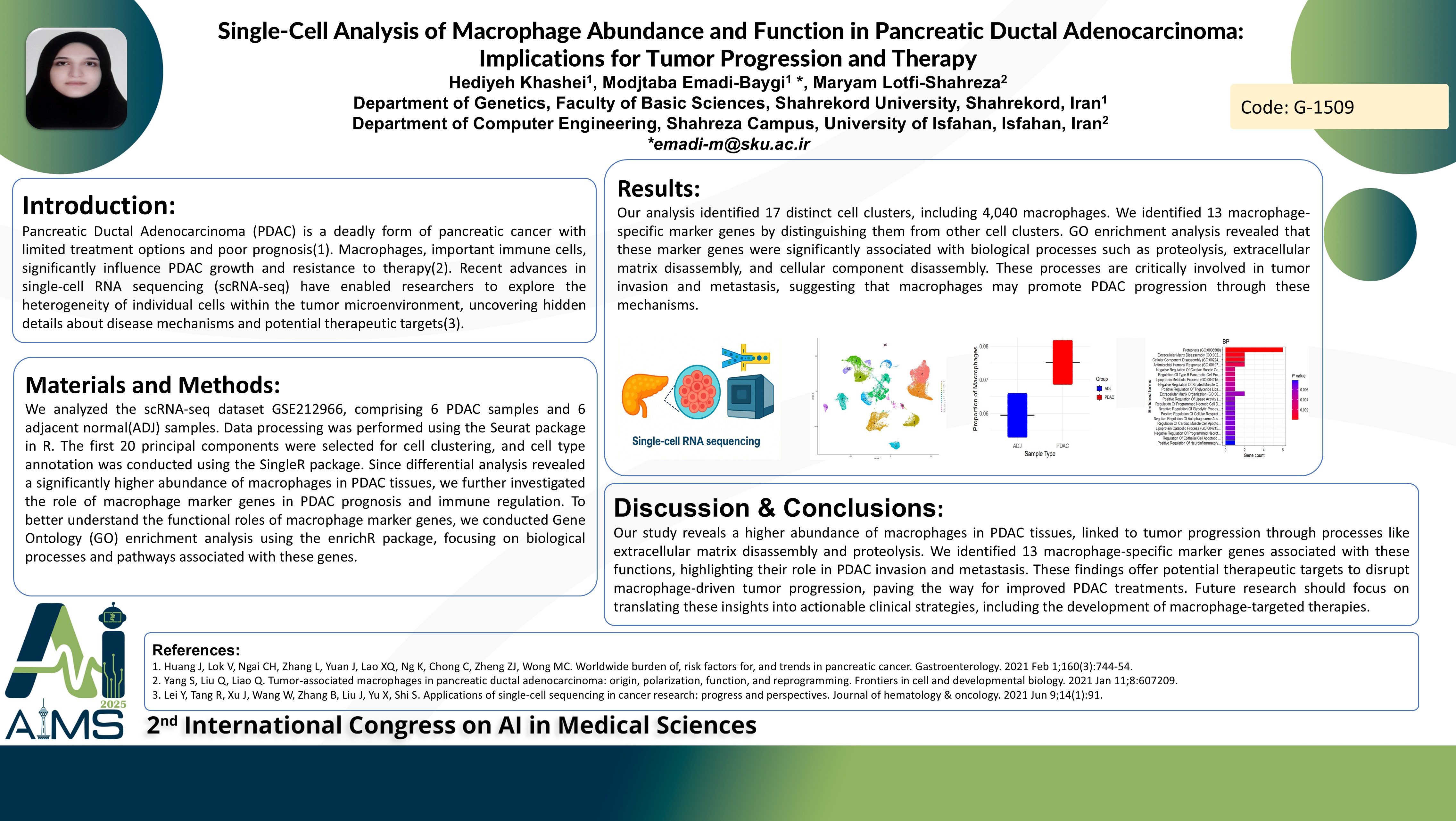
Background and Aims: Pancreatic Ductal Adenocarcinoma (PDAC) is a deadly form of pancreatic cancer with limited treatment options and poor prognosis. Macrophages, important immune cells, significantly influence PDAC growth and resistance to therapy. Recent advances in single-cell RNA sequencing (scRNA-seq) have enabled researchers to explore the heterogeneity of individual cells within the tumor microenvironment, uncovering hidden details about disease mechanisms and potential therapeutic targets. Methods: We analyzed the scRNA-seq dataset GSE212966, comprising 6 PDAC samples and 6 adjacent normal(ADJ) samples. Data processing was performed using the Seurat package in R. The first 20 principal components were selected for cell clustering, and cell type annotation was conducted using the SingleR package. Since differential analysis revealed a significantly higher abundance of macrophages in PDAC tissues, we further investigated the role of macrophage marker genes in PDAC prognosis and immune regulation. To better understand the functional roles of macrophage marker genes, we conducted Gene Ontology (GO) enrichment analysis using the enrichR package, focusing on biological processes and pathways associated with these genes. Results: Our analysis identified 17 distinct cell clusters, including 4,040 macrophages. We identified 13 macrophage-specific marker genes by distinguishing them from other cell clusters. GO enrichment analysis revealed that these marker genes were significantly associated with biological processes such as proteolysis, extracellular matrix disassembly, and cellular component disassembly. These processes are critically involved in tumor invasion and metastasis, suggesting that macrophages may promote PDAC progression through these mechanisms. Conclusion: Our study reveals a higher abundance of macrophages in PDAC tissues, linked to tumor progression through processes like extracellular matrix disassembly and proteolysis. We identified 13 macrophage-specific marker genes associated with these functions, highlighting their role in PDAC invasion and metastasis. These findings offer potential therapeutic targets to disrupt macrophage-driven tumor progression, paving the way for improved PDAC treatments. Future research should focus on translating these insights into actionable clinical strategies, including the development of macrophage-targeted therapies.
Keywords
Pancreatic Ductal Adenocarcinoma(PDAC), ScRNA-seq, Macrophage